This tutorial guide you how to use cosmo package to perform coarse-grained simulations of IDPs
Installation Cosmo
First, you need to follow the Readme file here to install the package.
Generate initial structure:
To generate initial structure, we can use a provided script: scripts/generate_structure_from_fasta/generate_structure.py
In this script, there are some parameters need to be changed:
seq = 'MRTQWPSPAKLNLFLYITGQRADGYHTLQTLFQFLDYGDTISIELRDDGDIRLLTPVEGVEHEDNLIVRAARLLMKTAADSGRLPTGSGANISIDKRLPMGGGLGGGSSNAATVLVALNHLWQCGLSMDELAEMGLTLGADVPVFVRGHAAFAEGVGEILTPVDPPEKWYLVAHPGVSIPTPVIFKDPELPRNTPKRSIETLLKCEFSNDCEVIARKRFREVDAVLSWLLEYAPSRLTGTGACVFAEFDTESEARQVLEQAPEWLNGFVAKGANLSPLHRAML'
protein_name = "2ww4"
# number of chains = nx*ny
nx = 2
ny = 2The long axis of molecule will be aligned along z-axis, we will expand the system along x-y plane.
- line 1: the fasta sequence of protein we are studying
- line 2: name of protein, will be used to write pdb output file name
- lines 3-4: number of copies in x-y dimension, in case simulation of multichains
Simulation control file:
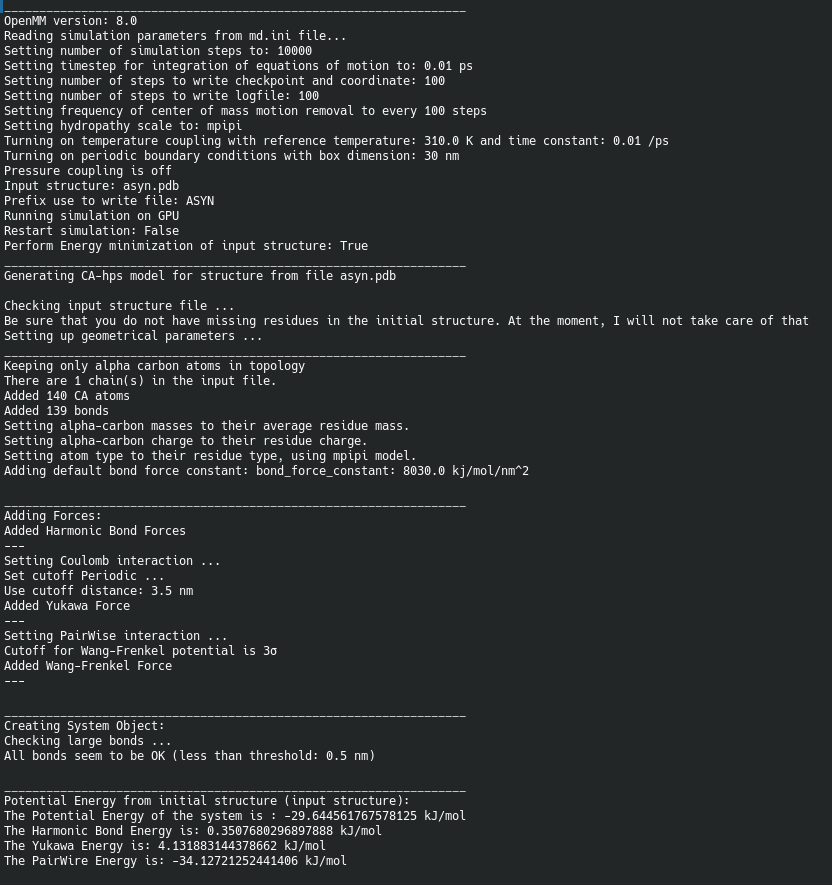
Simulation control parameters are saved in *ini file format (python file format): here, I will named it as md.ini
[OPTIONS]
md_steps = 10_000 # number of steps
dt = 0.01 ; time step in ps
nstxout = 100 ; number of steps to write checkpoint = nstxout
nstlog = 100 ; number of steps to print log
nstcomm = 100 ; frequency for center of mass motion removal
; select HPS model, available options: hps_kr, hps_urry, hps_ss, or mpipi
model = mpipi
; control temperature coupling
tcoupl = yes
ref_t = 310 ; Kelvin- reference temperature
tau_t = 0.01 ; ps^-1
;pressure coupling
pcoupl = no
ref_p = 1
frequency_p = 25
; Periodic boundary condition: if pcoupl is yes then pbc must be yes.
pbc = yes
; if pbc=yes, then use box_dimension option to specify box_dimension = x or [x, y, z], unit of nanometer
; otherwise, box_dimension is not required and does not take any effect.
box_dimension = 30 ; [30, 30, 60]
; input
protein_code = ASYN
pdb_file = asyn.pdb
; output
checkpoint = asyn.chk
;Use GPU/CPU
device = GPU
; If CPU is specified, then use ppn variable
ppn = 4
;Restart simulation
restart = no
minimize = yes ;if not restart, then minimize will be loaded, otherwise, minimize=FalseThis control file is almost self-explaination if you are familiar with other MD simulation packages.
Running simulation
To run simulation, there are two options:
Normal canonical simulation using Langevin dynamics:
hps-simulation -f md.iniIf you want to customize your simulation to use some advanced methods, make a copy of
examples/standard_example/run_simulation.pyCustomize the content of this script then execute the command:python run_simulation -f md.ini
Analyze simulation trajectory
Simulation will generate a psf file for topology and dcd file for trajectory, in addition, a log file that contains potential energy, kinetics energy, total energy and time remain to run simulations. But mainly you will need to analyze the trajectory to extract the information you need.
In the scripts/ directory, I provided some scripts I used in my study but you may need to develop for yours.